The human genome contains an estimated 19,000 genes. Those genes encode proteins that allow cells to carry out tasks such as ferrying oxygen molecules, fighting off diseases and communicating with fellow cells. But the function of most genes remains elusive, and scientists are still struggling to crack the human body’s full genetic code.
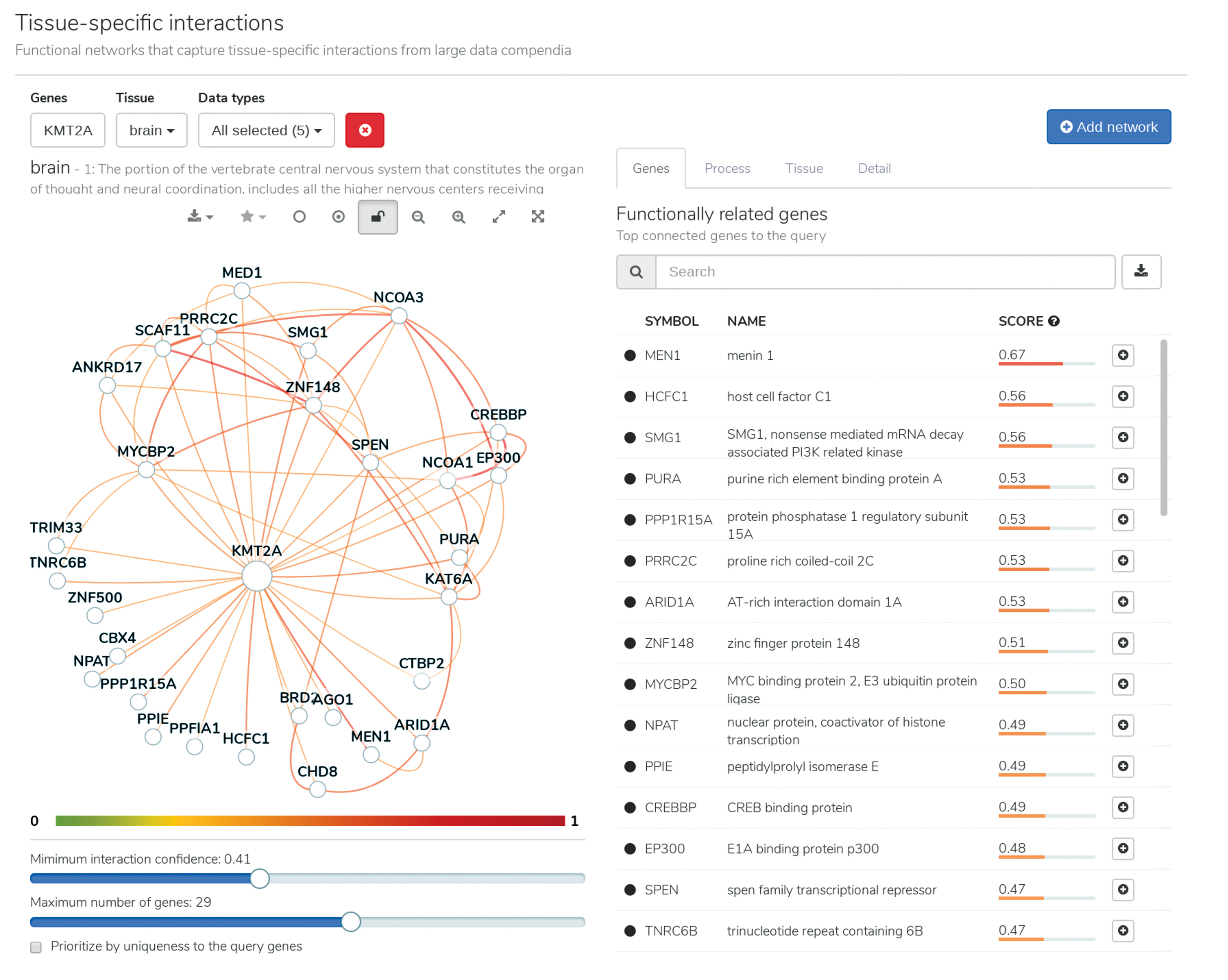
In March 2017, researchers at the Flatiron Institute’s Center for Computational Biology (CCB) soft-launched a new tool for decoding the human genome. The cloud-based software, called HumanBase, uses machine learning to trawl through decades of genomic research data for previously unseen potential biological connections. HumanBase can sleuth out how specific genes could potentially control cell functions, influence the expression of other genes, and contribute to disorders such as autism. Researchers can then carry out experiments that verify those potential connections.
“There’s a huge wealth of undiscovered knowledge in these data,” says Olga Troyanskaya, deputy director for genomics at CCB. “We wanted to build a single resource that could help biologists discover and leverage that knowledge.”
Historically, knowledge in the field of genomics largely rested in published findings and the heads of biologists. That’s changed, Troyanskaya says. New experiments now generate colossal datasets, but genetic associations are often too faint to uncover with any certainty from any one experimental dataset. Finding those connections requires looking at a much bigger picture, Troyanskaya says.
“A lot of these connections you just cannot see with traditional approaches,” she says. “You need computational algorithms that can pick up granules of data across multiple datasets. That’s impossible to do just in your head.”
“A lot of these connections you just cannot see with traditional approaches.”
HumanBase incorporates data from more than 38,000 genomic experiments and more than 14,000 scientific publications. The software standardizes all of those data before trained algorithms sift through the information looking for biological connections, particularly in the context of specific tissues, cell types and diseases.
HumanBase users can just type in a particular gene or disease and quickly receive a list of genes ripe for experimental scrutiny. For instance, if a gene often expresses alongside genes already associated with increased risk of Parkinson’s disease, that gene could be a tempting target for further research. “It’s guilt by association,” says project leader and CCB data scientist Aaron Wong.
The algorithms that power HumanBase have already proved their prowess. In August 2016, Troyanskaya, Wong and their colleagues reported online in Nature Neuroscience a substantial breakthrough in the hunt for genes associated with autism. Using a predecessor to the numerical tools employed in HumanBase, the researchers identified roughly 2,500 genes potentially linked to autism. Several of the most promising candidate genes had no prior genetic research tying them to autism. Scientists had previously identified 65 autism risk genes and predicted that 400 to 1,000 genes are likely involved in autism susceptibility.
HumanBase users are already tapping the software’s potential to generate new hypotheses and spark new experiments, but the software’s development is far from over. Troyanskaya, Wong and their team plan to add even more datasets to HumanBase’s knowledge bank every six months and to continue developing the algorithms powering the software. “We want to build something that biomedical scientists can rely on,” Wong says. “We want them to incorporate HumanBase into their research workflow — to drive new hypotheses and follow-up experiments.”




