New Model Predicts How Bacteria Navigate Obstacles to Spread

Trying to predict how bacteria will spread is like predicting the flight of a leaf in a windstorm — it’s a complicated and chaotic business. Factor in bacteria’s encounters with objects such as corners or surfaces and the calculation gets even more complex. While such predictions are incredibly difficult, they’re also vital to reining in the spread of bad bacteria (such as infections) or getting good bacteria to the places we want them to go (such as bacteria engineered to deliver drugs to tumors).
In a new study published March 21 in Proceedings of the National Academy of Sciences (PNAS), Henry Mattingly of the Simons Foundation’s Flatiron Institute presents a new computational method for predicting how bacteria navigate obstacle-filled settings. His model is the first to theoretically predict how bacteria move in complex environments.
“There’s great interest in how bacteria spread because they can play a role in everything from infections and agriculture to designing better vehicles for drug delivery and environmental waste clearance,” says Mattingly, an associate research scientist at the institute’s Center for Computational Biology (CCB). “With this model, we have a method for determining how bacteria will spread, even when they encounter obstacles along the way.”
Bacteria aren’t the most efficient or predictable organisms when it comes to navigating the world around them. This work focuses on bacteria that swim through liquid environments, driven by propeller-like tails and changing direction completely at random. As it turns out, a key determinant of their travel is how often they reorient themselves, with too many or too few direction changes slowing them down.
If bacteria are in an environment that includes obstacles, which is often the case, they can easily become slowed or trapped. When a bacterium hits a roadblock, it can either slide along the surface and continue moving, much like a skateboarder on a rail, or become trapped in a corner and repeatedly reorient itself until it finds a way out, like a Roomba at the edge of a room.
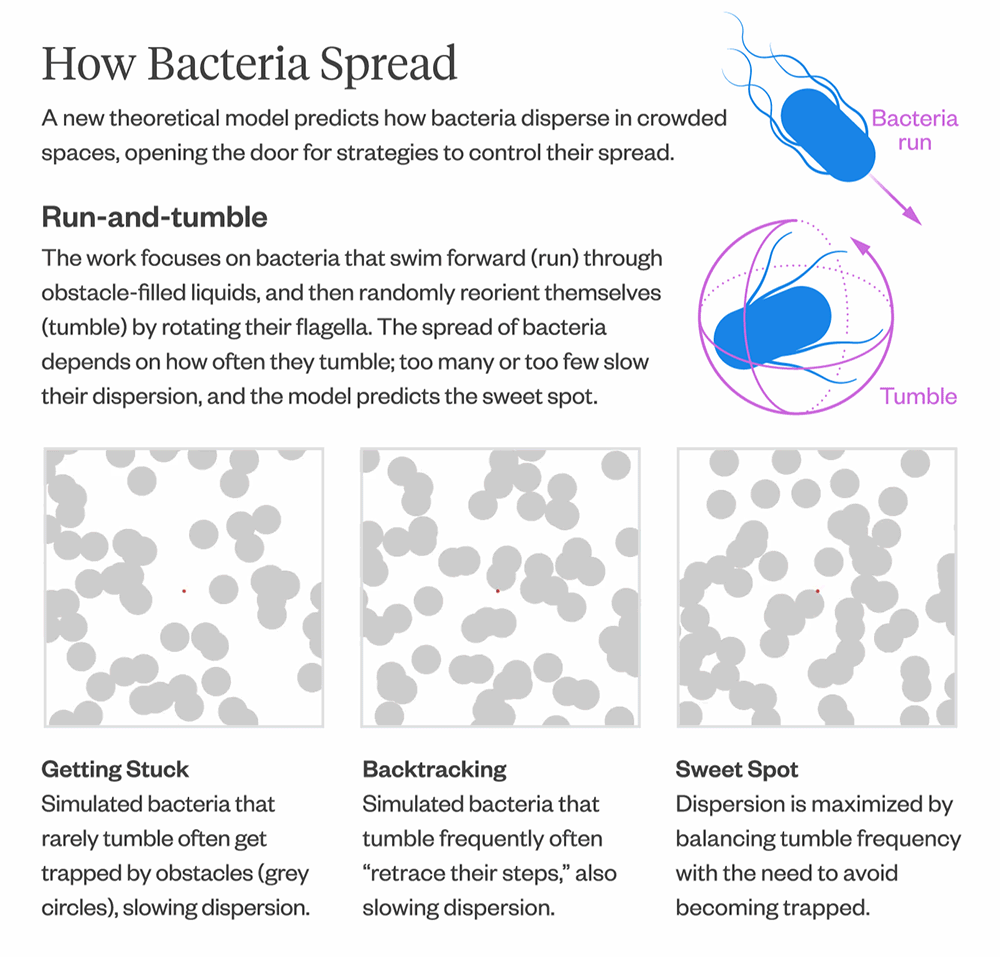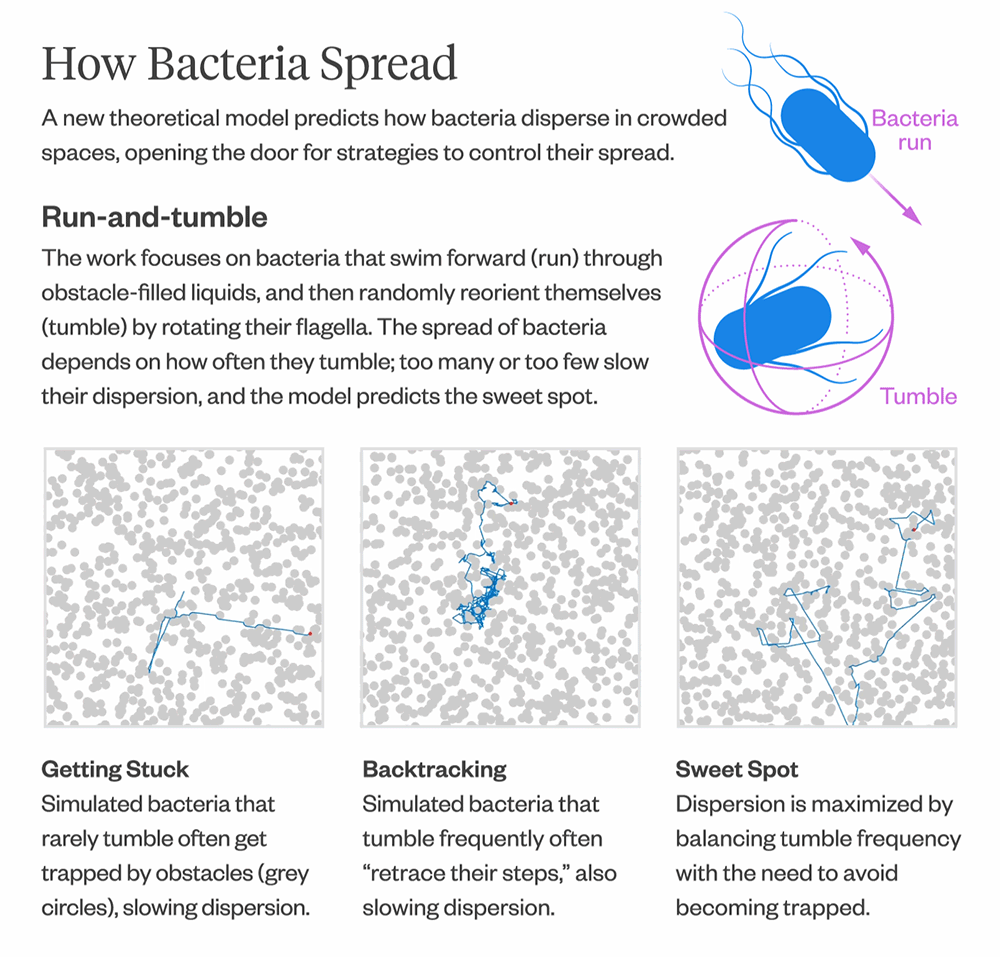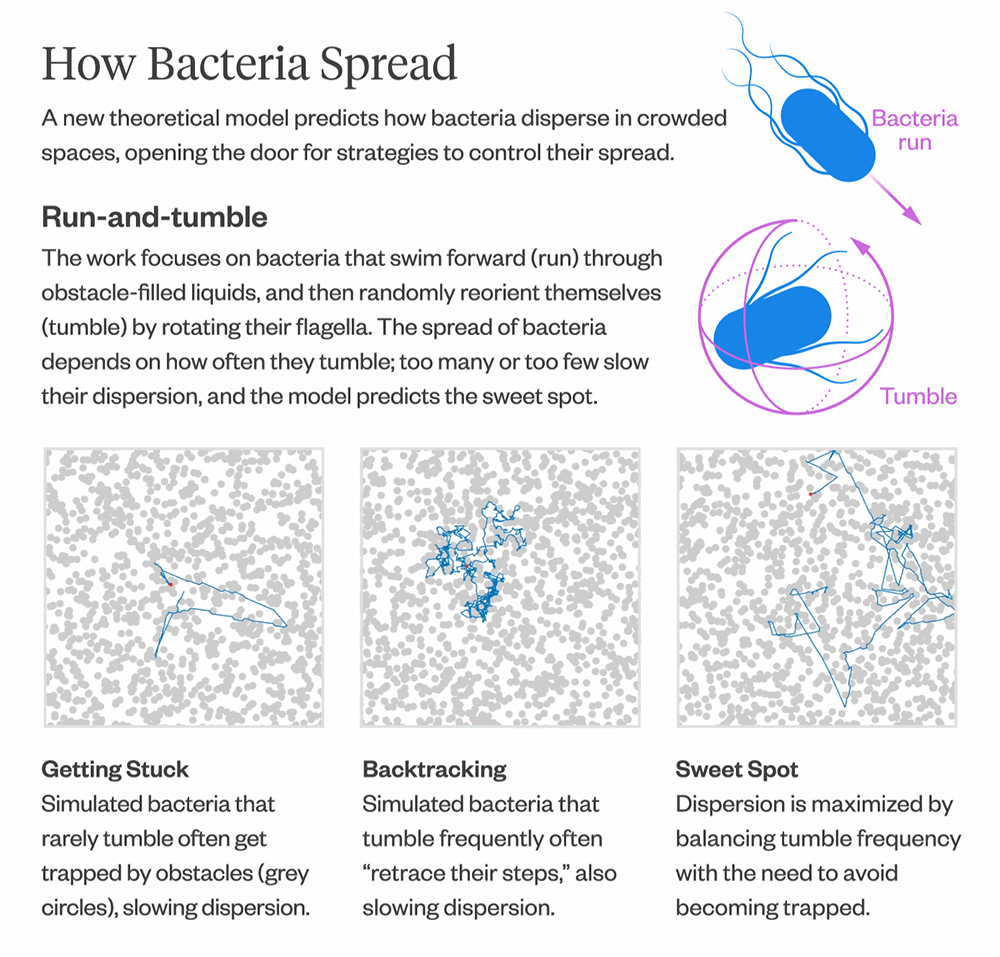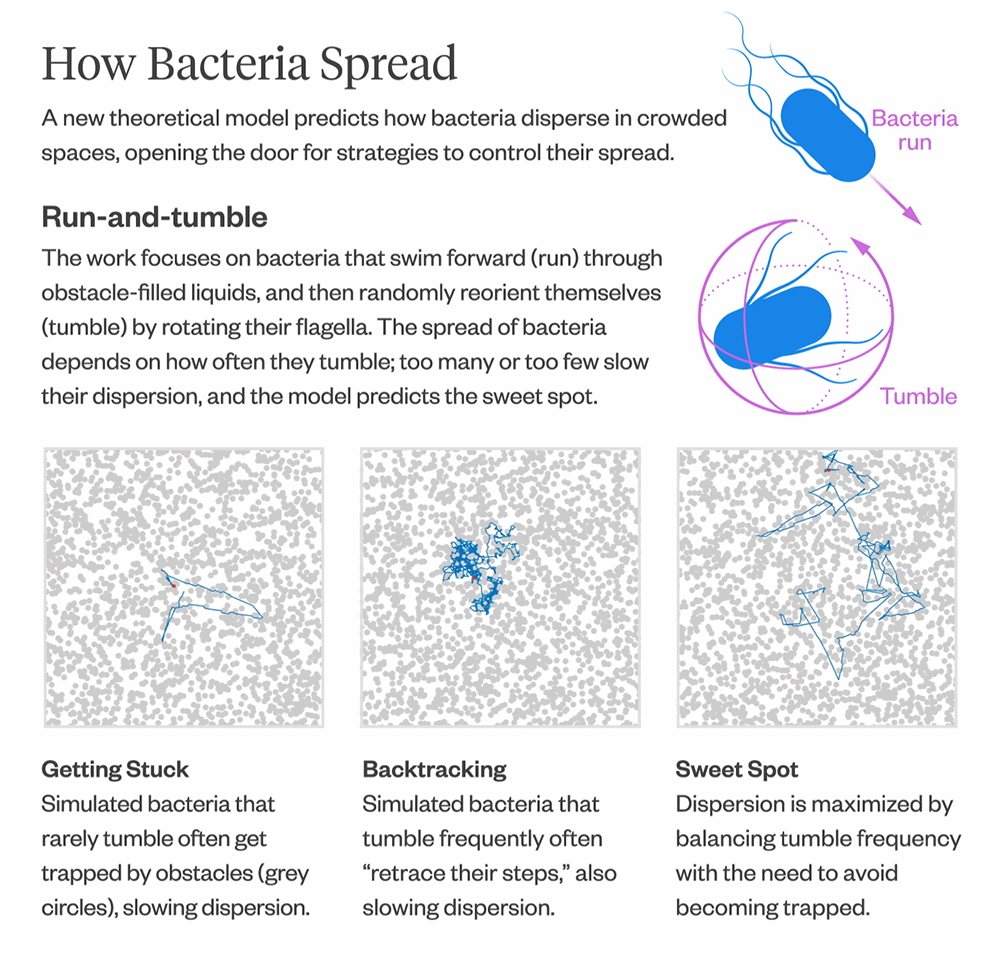
There have been many simulation studies over the years that have modeled bacterial movement in crowded environments, but there hasn’t been a theoretical model for predicting how fast or slow bacteria spread.
“Analytical theories of this process have been lacking because disordered environments are complicated. The obstacles may be arranged randomly, and bacteria interact with obstacles in complex ways,” says Mattingly. “We haven’t had a robust understanding of how properties of the environment and cell motility influence bacterial dispersion.”
Mattingly decided to focus his model on ‘surface states’ that describe where bacteria are in relation to obstacles (or surfaces) in the environment. He defined three basic surface states for a bacterium: moving through the environment uninterrupted, encountering an obstacle and sliding along it, and getting stuck in a corner.
“Boiling it down to these three surface states was a very optimistic place to start, but it ended up working well and still being the core of the method, in the end,” says Mattingly.
By deriving how often a bacterium transitions between surface states, Mattingly could determine the probability of its being in each state. This depended on how obstructed the space was and how often the bacterium changed direction. By adapting mathematical techniques previously developed for diffusion of passive substances like dyes, he was able to construct a model to predict bacterial diffusion.
Then it was time to put the model to the test. Mattingly coded simulations of swimming bacteria, varying the number of obstacles (represented as randomly placed circles) and the frequency at which bacteria changed directions. He then watched the bacteria disperse and compared their diffusion to the calculations.
“[The model] predicted diffusion really well,” he says. “It’s very surprising that bacterial motion in this complicated environment can be compressed down to just these three states.”
The model also provides insights into how bacteria optimize their diffusion. Bacteria that infrequently change direction are more likely to get trapped in corners and remain there for longer, resulting in slower diffusion. Conversely, bacteria that change direction frequently tend to “retrace their steps,” which also leads to slower diffusion. The model can help predict the “sweet spot” between the two extremes, where bacteria maximize their diffusion by balancing the frequency of direction changes with the need to escape from corners.
Knowing this will make it easier for scientists to design lab-made bacteria-driven microparticles that target cells for applications such as drug delivery.
“If you’re designing a microswimmer, you’ll want to know the optimal rate at which bacteria should reorient to keep moving outward but also get out of traps quickly,” says Mattingly. “This model can help identify that rate based on the number of obstructions in the environment.”
Next, Mattingly wants to test the model on real-life bacteria.
“I’d like to work with somebody who does experiments with real bacteria,” says Mattingly. “I’m sure there are aspects of a real system that weren’t included in the model, but the hope is that a similar approach with the right ingredients will describe bacterial dispersion in experiments.”
Information for Press
For more information, please contact Stacey Greenebaum at press@simonsfoundation.org.


