
Exponentially Important: The Scientific Origins of PCR
In the age of COVID-19, we all know the drill. Even if we haven’t done it ourselves, we’ve heard of others going to a clinic or a drive-through site for virus testing—offering a nostril or throat for a quick, confident swab by a PPE-clad health-care professional. What we rarely think about is what happens afterward. Using the sample on the tip of the swab to identify the presence (or absence) of the infinitesimally small SARS-CoV-2, the strain of coronavirus that causes COVID-19, is like trying to find a needle in a haystack—from the vantage point of the moon. And yet we are able to do it. Thanks to laboratory techniques like PCR—polymerase chain reaction, a simple method for “amplifying” (copying) tiny segments of DNA until they are numerous enough to identify—we are able to detect the presence of this dangerous, minuscule virus with laser-sharp precision.

Considered the gold standard of COVID-19 testing for its accuracy and reliability, PCR can extract, amplify, and zero in on the exact genetic code of SARS-CoV-2. People around the globe have looked to PCR tests for dependability as we scramble to contain a coronavirus pandemic that has brought the world to its knees. But what many people don’t know is that the invention of PCR in the 1980s was a scientific turning point with momentous consequences. Its discovery came on the heels of decades of advances in basic science research, thanks to support from government agencies, industry, and private philanthropy. Now ubiquitous in both research and clinical laboratories, with a range of applications from molecular biology to genomics and even forensics, PCR made entire fields of scientific work easier and fueled the growth of the biotech industry.
“PCR is in one sense an evolutionary shift,” explains Richard Doyle, PhD, a historian of science at Pennsylvania State University. American biologist James Watson, PhD, and English physicist Francis Crick, PhD, set that shift in motion when they modeled the double-helix structure of DNA in 1953. “With PCR 30 years later,” continues Doyle, “you could actually manipulate this DNA. The chain reaction itself then causes a chain reaction. You get the whole ‘Boom!’ effect. Four molecular biologists and a computer can start a company. You get massive shifts in drug design and Big Pharma—all because you can directly extract genetic information and manipulate it, which is ordinarily the job of evolution through organisms.”
Ever since Kary Mullis, PhD, won a share of the Nobel Prize in Chemistry in 1993 for the invention of PCR, the scientific origin story of the polymerase chain reaction has been a tale of imagination and vision. Yet if we look closer, it’s really a story of teamwork and collaboration that could never have unfolded without the efforts of many hard-working scientists. And it’s a perfect example of how basic science research, from bacteriology and chemistry to human genomics and beyond, opens the door to advances that can change the course of science, help fight pandemics, and hold the answers to questions we have yet to ask.
Prequels to PCR
Mullis is often cited as the author of a process that revolutionized science (much has also been written about his sometimes difficult personality, affinity for psychedelic drugs, and belief in astrology—factors that some say made him an unlikely Nobel laureate). But his work piggybacked on that of many others who came before him.
Much of the work that laid the foundation for the invention of PCR was based in bacteriology and focused on some of the tiniest life forms. “Nearly 100 years of research using bacteria has helped us figure out many things related to DNA,” says Jade Wang, PhD, a bacteriologist at the University of Wisconsin, Madison. “If you started only from higher organisms, we would never have reached the understanding we have now. It’s also much cheaper—hundreds, thousands, or maybe tens of thousands times cheaper—to do [basic research] in model bacteria.”
A few years after Watson and Crick (thanks to X-ray crystallography images provided by chemist Rosalind Franklin, PhD) unlocked the double-helix structure of DNA in 1953, bacteriologist Arthur Kornberg, MD, advanced the study of DNA replication when he and colleagues discovered the first DNA polymerase, in the bacterium Escherichia coli (E. coli). DNA polymerases are enzymes that catalyze the synthesis of new copies of DNA, and their discovery was essential for understanding how DNA replicates itself to repair and maintain the genetic material in all living organisms.
Kornberg, who won a 1959 Nobel Prize for this discovery, also hypothesized the process of DNA replication—but it was geneticist Matthew Meselson, PhD, and biologist Franklin Stahl, PhD, who successfully demonstrated it, also using E. coli, in the Meselson-Stahl experiment. “Some people say this is the most elegant molecular genetic experiment,” Wang notes. “[It] showed that DNA is replicated by a semiconservative mechanism, meaning that each strand of DNA serves as a template for the production of a new strand.” It was this very mechanism that PCR would leverage to amplify genes.
Research into the genetic code—the key to our understanding of all life forms—was well underway. So was a new age of synthetic biology. In 1971, biochemist H. Gobind Khorana, PhD, and colleagues offered the first description of “repair replication” of short, synthetic DNA strands—which involved bracketing a targeted DNA sequence with a pair of oligonucleotides, then copying the sequence using DNA polymerase. This work was funded by the federal government, universities, and the Life Insurance Medical Research Fund. In their paper, they described basic principles of PCR—the use of primers facing each other, the addition of DNA polymerase for replication, and the reiteration—but stopped short of demonstrating the groundbreaking process.”

Meanwhile, in the wilds of Yellowstone National Park in the late 1960s, microbiologist Thomas Brock, PhD, discovered a species of bacterium, Thermus aquaticus (T. aquaticus), which thrived in the superheated environs of the park’s thermal springs. “Some people called [basic research] useless because it had no practical aims… I think the evidence is that basic research is what practical aims build on,” Brock said in his acceptance speech for an honorary degree from from the University of Wisconsin-Madison. Yet his research carried enormous import for understanding how life can exist at high temperatures. It would also be key to the development of PCR—because T. aquaticus produces a heat-stable DNA polymerase, called Taq polymerase, that would make PCR more practical and automatable. (Today, in addition to Taq polymerase, PCR uses many different heat-stable enzymes—such as Pfu polymerase from Pyrococcus furiosus, Tli polymerase from Thermococcus litoralis, and Tth polymerase from Thermus thermophilus—all obtained from thermophilic organisms through basic science research.)
Finally, in 1977, biochemist Frederick Sanger, PhD—working with a bacteriophage (a single-stranded DNA virus) that affects E. coli—came out with a new, rapid way to sequence DNA. This work not only won Sanger a Nobel Prize in 1980 (he’d also won it in 1958, for determining the structure of insulin), but it helped to plant a seed in Mullis’s mind and make PCR easier to achieve.
Mullis’s “aha moment”
Though all the pieces were in place for the invention of PCR, it wasn’t until Mullis put them together that the idea began to crystallize. The year was 1983, and Mullis was driving on a moonlit April night from the Bay Area—where he worked with oligonucleotides at the biotech company Cetus Corporation—to his weekend cabin in northern California’s redwood country. While a chemist friend napped in the passenger seat, Mullis was planning a DNA-sequencing experiment in his head when he stumbled upon a thought so startling that he stopped the car, twice, to scribble notes. What if, under the right conditions, the DNA fragments he envisioned could reproduce themselves in a reiterative fashion?
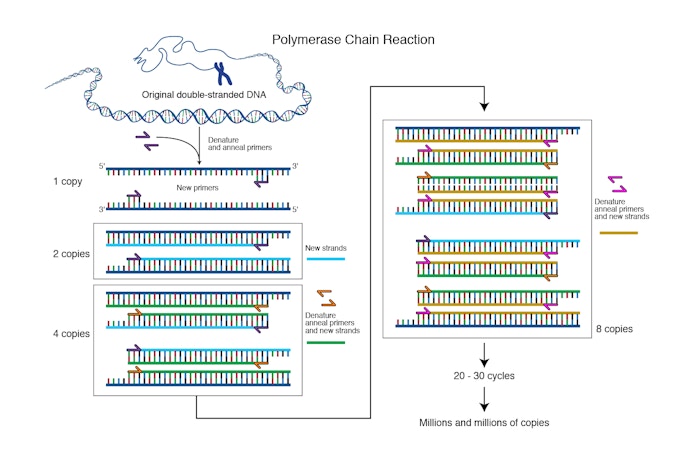
“From the glove compartment I pulled a pencil and paper . . . and started drawing lines of DNA molecules hybridizing and extending, the products of one cycle becoming the templates for the next in a chain reaction,” recalled Mullis in a 1990 Scientific American essay. The basic process of PCR was unfolding in his head. Using oligonucleotides in the manner that Khorana had demonstrated, he would create a DNA fragment. Then, by adding a polymerase reagent to the fragment, he could create replica DNA strands exponentially. One strand could become 2, then 4, 8, 16, 32, 64, and so on, leading to over 1 million copies in a matter of minutes. Using readily available enzymes in a lab, he could possibly make unlimited numbers of copies of genes. “It was difficult for me to sleep that night,” he said, “with deoxyribonuclear bombs exploding in my brain.”
Mullis suspected he was on to something and spent the next few months designing and preparing for an experiment to show proof that PCR can work. “It was a nice, simple idea,” says Henry Erlich, PhD, who was then head of human genetics in Cetus and is now a senior scientist at Children’s Hospital Oakland Research Institute. “The issue was, could you actually get it to work?” When he was ready, Mullis ran his experiment and found that it does.
Lighting up the right gene
The use of PCR then extended into the work of Erlich. At the time, Erlich and his colleagues were trying to develop a prenatal diagnostic test for sickle cell anemia, which involved looking for mutations in the gene for beta globin. To see if they could use PCR to amplify the beta globin gene, they split up into groups—Mullis working with his technician Fred Faloona and Erlich working with Randall Saiki, with additional support from Norman Arnheim, PhD; Steve Scharf; Glenn T. Horn; and Thomas White, PhD, head of research at Cetus.
While Erlich and the others worked with human cells to try to amplify the beta globin gene, Mullis and Faloona tried a simpler method, looking for the gene in a circular piece of cloned DNA. Yet they couldn’t get the experiment to work; instead of amplifying the specific gene they wanted, they amplified many DNA segments. In those days, researchers used radioactive probes to “light up” the specific gene segments they were looking for (today, they use fluorescent probes). It was Saiki who finally succeeded, after months of trial and error, at amplifying and lighting up the beta globin gene.
“When you’re trying to work out something, you try a lot of different conditions—different temperatures, different concentrations of the primers, different concentrations of the DNA polymerase, different concentrations of the nucleoside triphosphates,” explains Erlich. “[Saiki] actually found one set of conditions where he could see a specific band, a 110–base-pair fragment. You synthesize a lot of stuff, but about 1% of it was actually beta globin, and [with the probe] you could see that—bam!” It was a eureka moment. The team published the first study to reference PCR in 1985, followed in 1989 by Mullis and Faloona’s landmark paper about PCR.

The “revolution within the [PCR] revolution,” says Erlich, came when David Gelfand, PhD, and Susanne Stoffel succeeded at isolating and purifying native Taq polymerase—the heat-resistant enzyme from T. aquaticus—making it no longer necessary to add more enzymes after each cycle. After that, PCR use exploded. Cetus partnered with lab instrument maker Perkin-Elmer to develop a commercially available thermal cycler (aka PCR machine), which would soon become ubiquitous in laboratories. In 1987, PCR was used to detect HIV in cell lines. And the forensics field quickly picked up on PCR’s ability to identify an individual’s DNA from a single drop of dried blood or semen or a single strand of hair from a crime scene.
A variation of PCR (called multiplex PCR) was developed in 1988 where more than one DNA template can be amplified in a single reaction tube. This method was later applied to analyzing ancient DNA. PCR was also key to the success of the Human Genome Project (HGP), launched in 1990 and completed in 2003, with its grand goal of sequencing and mapping the entire human genome. The HGP led to the creation of the Broad Institute of MIT and Harvard—a biomedical research consortium funded by philanthropists Eli and Edythe Broad—which has served as a major PCR diagnostic testing lab during the COVID-19 pandemic.
Scientists continued to refine PCR yet further, creating even more applications and variants for the process. These tools would go on to support basic science research and become the bread and butter of laboratory work, in health care and beyond.
Fluorescence and real-time PCR
One major variant of PCR—which would be key to developing highly specific diagnostic tests like the assay for SARS-CoV-2—was real-time PCR, designed by Cetus scientists Russell Higuchi, PhD, Bob Griffith, and others in the early 1990s. Higuchi and Griffith stumbled on this variant serendipitously, while trying out different conditions that would allow them to visualize the products of PCR without opening their test tubes. One idea was to add ethidium bromide, a fluorescent reporter dye, to the completed PCR—but Griffith accidentally added it at the beginning of the experiment. To their surprise, PCR not only still worked in the presence of the dye, but the dye also allowed them to view the creation of DNA copies in real time, as the fluorescence signal increased with each PCR cycle. In other words, the fluorescent dyes lit up as they bound to the target DNA, allowing scientists, for the first time, to quantify the amount of DNA over time. Hence real-time PCR is also known as quantitative PCR (or qPCR).
The creation of this particular PCR variant would not have been possible without basic science research into organic fluorescent compounds. Over the course of the 19th century, British astronomer Frederick Herschel provided one of the first descriptions of the phenomenon of fluorescence, British physicist George Stokes described fluorescence in greater detail, and research in Germany led to the development of the first fluorescent microscopes. In 1942, fluorescent antibody labeling was developed, aided by a grant from the International Cancer Research Foundation and other philanthropic grants. Basic science research into the characteristics of green fluorescent protein led to its widespread use as a biomarker in research.
When fluorescent markers replaced radioactive probes in PCR technology, scientists had a safer way to amplify DNA. The development also gave them more data, because now they could measure the fluorescent signal in real time, with mathematical precision. To analyze these data, scientists use mathematical models and growth curves to determine the quantity of DNA present. This opened the door to new applications—such as measuring viral load or quantitating gene expression. “The signal that’s generated during [qPCR] is a curve,” explains John Moskow, PhD, of MilliporeSigma, a life science company that provides scientists with lab materials and technologies, including PCR assay kits. “By using mathematical models, you can come up with the best way to identify the unique point on the curve for each [DNA] sample and, hence, get an exact number.” Although most PCR assays for SARS-CoV-2 do not measure viral load but simply report the presence or absence of the virus, laboratories still use real-time PCR/qPCR for its sensitivity, specificity, and speed, which make it the gold standard for such testing.
Not to be confused with real-time PCR, another variant of the process, called reverse transcription PCR (RT-PCR), allows researchers to amplify genetic material from RNA. RT-PCR involves adding another enzyme—called reverse transcriptase—to the PCR process. Reverse transcriptase copies the RNA strand into a complementary DNA strand, which can then be amplified. Among other applications, RT-PCR makes it possible to diagnose HIV and SARS-CoV-2, as they are both single-stranded RNA viruses. In assays for COVID-19, labs use a combination of RT-PCR and qPCR (known as RT-qPCR) to make copies of any viral RNA present, so the virus can be detected.
PCR for the pandemic era
Basic science research aimed at further refining PCR continues to this day, given its promise of helping to fight not just COVID-19 but also future pandemics. In chemistry labs, such research is developing ultrasensitive fluorescent dyes. Scientists also continue to investigate polymerase enzymes and their buffer systems (that is, their chemical environments) to find those that amplify DNA most efficiently. Such work has been essential to revolutionizing the manufacture of ready-to-use PCR kits, which come with swabs and polymerases for fast virus testing, among other uses. Also in development are refinements of isothermal PCR—a variant of PCR that doesn’t require a thermal cycler machine, making it ideal for field diagnostics in areas far from a hospital or clinic.
Perhaps the next generation of PCR will be digital PCR, which apportions the sample among tens of thousands to hundreds of thousands of droplets for PCR analysis. The droplets are so tiny that they will contain at most one copy and perhaps none of the sequence being sought. “You analyze each droplet separately . . . and you count the number, as opposed to getting a logarithmic curve,” explains Moskow of MilliporeSigma. “It tends to have better sensitivity. It also has the inherent ability of being internally quantitative, meaning that you can count the absolute number of molecules that were in the starting material. That’s probably the next 20 years of PCR—building out the digital piece.”
The science around PCR makes it clear that knowledge generation is a long game, and when pandemics like COVID-19 strike, the stakes couldn’t be higher. “You have to grow a culture of knowledge . . . and that requires a long-term investment,” says Doyle, the Penn State historian of science. “Without basic science, not only would we not come up with PCR, but we wouldn’t know how to train people to do the [COVID-19] test, and we wouldn’t know how to teach people and policy-makers how to interpret it. Minimizing suffering and maximizing progress means that you have to commit yourself to knowledge. Philanthropists are a crucial part of that ecosystem.”
About the Author
Wendy Kagan is a writer, researcher, and editor based in New York’s Hudson Valley with expertise in science-based health and wellness.
This article first appeared as part of the Science Philanthropy Alliance‘s COVID-19 Basic Science Prequels series. Decades before COVID-19 gripped the world, basic science researchers were laying the groundwork for lifesaving medical and technological breakthroughs now central to the world’s pandemic response. With support from the Kavli Foundation and the Simons Foundation, the Science Philanthropy Alliance has enlisted a team of science writers to explore these science origin stories. Each month, the COVID-19 Basic Science Prequels will unpack the people, history, and serendipitous discovery behind topics that now dominate our daily lives.