The Connected Connectome Part 2: Beyond the Fly
An old saying in neuroscience goes: “Neurons that wire together fire together.” In other words, the connections between neurons govern the brain’s activity, controlling everything from physical movement to consciousness.
Or so we think. Decades of experiments in animals show neurons building and breaking connections as an animal learns, but what those connections actually mean largely remains a mystery. In recent months, reconstructions of the connections in the fly brain have already rewritten much of what neuroscientists thought they knew about this circuitry. “The general fundamental question is: How does the physical brain relate to what the brain does?” says Jeff Lichtman, a neuroscientist at Harvard University. “We know so little about the physical brain we can’t even ask that question in a scientific way.”
Building off recent successes in the fly, a map of the mouse brain connectome — the wiring diagram of its 70 million neurons — could start to provide this physical picture, and the path to that goal is now starting to take shape. The MouseLight Project at the Howard Hughes Medical Institute’s Janelia Research Campus, for instance, is mapping connections between neurons across the brain, while an initiative funded by the Intelligence Advanced Research Projects Activity (IARPA) is trying to reconstruct all the connections in a cubic millimeter of a rodent’s brain.
Such projects can provide a rough idea of how the brain’s neurons and regions communicate with one another, setting some ground rules for how a neuron’s shape regulates its function, or which brain regions are involved in a circuit of neurons. But to truly understand how emergent properties like memory and vision arise from a loop of connected cells, researchers need to dig deeper into the mess of tiny branches called axons and dendrites that project from neurons. They need to peer inside the 15-nanometer-wide junctions between neurons to see which cells share a close relationship — and which are key hubs in the complex neuronal web that makes up the brain.
The ability to physically see wiring at this scale may challenge assumptions and theories that form the bedrock of neuroscience. “There are hints, bits and pieces, that there’s something very interesting going on,” says Karel Svoboda, a neuroscientist at Janelia. “Only after having all of the neurons and chains of connections can you get at the logic of this network. [Electron microscopy] is the only game in town for how to get at that.”
Electron microscopy, or EM, which creates images with nanometer-scale resolution, has already proved itself in the fly connectome (see part 1 of “The Connected Connectome”). In September, a team of 108 scientists published a paper detailing a project called FlyEM, the most complete map of the wiring diagram in the Drosophila brain. The resulting database includes 25,000 neurons in the fly brain and the 20 million connections, or synapses, between them. FlyEM, which launched online earlier this year, has been transformative for the field, says Clay Reid, a neuroscientist at the Allen Institute for Brain Science. “I don’t think anyone does work in Drosophila without it.”
But compared to imaging the fly brain, imaging a mouse connectome at this level presents a daunting task. The entire fly brain is about the same size as one mouse cortical pyramidal cell from the tip of the axon to the tip of a dendrite. And mice are not bigger flies: To study mammalian brains, researchers have to first work out the mundane details of how tissue is stained, cut and scanned on an electron microscope. Tasks like data processing, training automated machine learning to identify neuronal connections, and programming the technology that stitches the images together are already challenging with flies, but their complexity increases exponentially with larger animals.

Big neuroscience
In a September commentary in Cell, about two dozen authors detailed the challenges of building a mouse connectome while calling for the resources to build it. “It’s too big a project for any individual to take on,” Lichtman says. Rather, the project will require collaboration on an order rarely seen in neuroscience — making it more akin to the search for the Higgs boson than the study of a model organism.
One obvious source of funding would be the National Institutes of Health’s BRAIN Initiative. In 2019, the agency decided that over the next five years, BRAIN would fund several large projects rather than doling out money to smaller ones. Building the mouse connectome ranks high on the initiative’s list of possible flagship projects. “These could truly transform the way we do neuroscience,” says BRAIN Initiative director John Ngai, “not just research, but also cures.” Researchers from all subfields of neuroscience could use the connectome to begin asking questions about the basic principles that govern brain function across species.
Beginning this year, the NIH is hosting a series of mouse connectome workshops to help set priorities for the types of new technologies to pursue, the type of mouse brain to reconstruct, and the questions that the connectome would answer. “We first need to understand what needs to be scaled and then how to scale it,” Ngai says.
The workshops are cohosted by the U.S. Department of Energy (DOE), whose computing power was crucial to the completion of the Human Genome Project. Processing the estimated 1 million terabytes of data in a mouse connectome would be orders of magnitude more complex than mapping the human genome, whose sequence comprises just a few hundred megabytes by comparison. “It’s a great challenge for supercomputing — that’s what makes it exciting,” says Sebastian Seung, a neuroscientist at Princeton University. “You don’t want to build a supercomputer and let it sit there. You need to find problems at the frontier.”
The neuroscience field has nothing like DOE national laboratories, which include such powerhouses as Argonne and Oak Ridge and serve as home bases for scientists from all over the world to collaborate on massive physics experiments. By creating such a central hub for connectomics, the NIH can further bolster crucial cross-disciplinary thinking from all corners of neuroscience. For example, the hub could include improved experimental resources like multiple electron microscopes in one place, allowing researchers to churn through data quickly, plus unparalleled computing power to run projects like machine learning algorithms that identify synapses and stitch images together.
But even if the mouse connectome project is funded, Ngai estimates it will take at least five years before technology catches up enough to allow scientists to actually start constructing the map. With current technology, he says, “I think we could grind through a whole mouse brain in some way, shape or form, but it would probably take us a decade or two.”
Devil in the details
That said, in some ways, imaging the mouse connectome may be simpler than it was for the fly, says Ken Hayworth, a neuroscientist at Janelia. While fly axons and synapses are a mess of fine tangles, those same structures are much clearer and easier to identify in mice; it is the difference between untangling the strands of a cotton ball and a bundle of electrical cables. And unlike insect neurons, mammalian axons are sheathed in myelin, a protein that readily picks up stains and allows researchers to see the edges of the wires.
But the mouse presents a score of new technological hurdles, each less sexy than the last. The procedure for staining a mouse brain, for instance, is onerous and expensive, using heavy metals like uranium, lead and osmium to darken the membranes of neurons. It’s difficult to know whether the stain has penetrated into the center of the mouse’s brain — so difficult that Lichtman’s group has been forced to use X-rays to see how deeply the brain has absorbed the metals.
Cutting mouse brains into slices thin enough to image on a microscope is also a surprisingly hard problem. Several groups are using a knife edged with diamond — the only material that can cut through the metal-stained tissue — to produce slices 30 nanometers thick. These thin sections tend to fold up and tear easily, making them extremely difficult to work with.
Researchers at Janelia are addressing that problem by working backward. After embedding a brain in a block of plastic, they use a scanning electron microscope (SEM) to image the exposed portion of the brain. Then they slice 30 nanometers off the block and the brain to expose a new surface to image. Eventually, they can reconstruct a 3D structure from the series of images.
But the diamond knives are only 4 millimeters long, far from long enough to cut through a 10-millimeter-thick(CK) mouse brain. Cutting a brain into usable chunks destroys tissue along the edges of the cuts, and each slice of a diamond knife destroys 7 nanometers of tissue. That’s a lot of missing data for researchers trying to reconstruct synapses only a few nanometers wide. “Any little blemish in data can screw you up,” Svoboda says. “If you have these vast datasets, you have to be nearly perfect.”
Winfried Denk, a physicist at the Max Planck Institute of Neurobiology, agrees, particularly when it comes to imaging. “It has to work 9,000 out of 9,000 times,” he says. “That’s reliability more than is typically known in a research lab.”
But reconstructing the connectome will require the imaging to be not only good, but fast. Right now, Denk says, it would take at least three years to image one mouse brain — if everything went perfectly the first time. But he thinks that several steps can be shortened. For instance, a more intense and precise stain would mean that computers wouldn’t have to spend time picking the signal out of the noise. Denk’s group is also developing a way to divide the plastic block in half and image the exposed tissue on one side while the machine carves a 30-nanometer slice off the other half, then switch sides. That would halve the time needed to image the brain. “This is a transition for scientists because we have to think more like engineers,” Denk says.
When images are pieced back together, a few missing nanometers can make it hard to tell which axon is which. “It’s really easy to jump from one axon onto its neighbor and end up in another part of the brain,” says Kristen Harris, a neuroscientist at the University of Texas at Austin. “It’d be a catastrophic failure.”
Harris’ group is developing a microscope that can image slices at 1-nanometer resolution, which would allow researchers to see structures like the signaling vesicles in synapses. The system, called Tomo-SEM, works like a hospital’s CT scan: imaging virtual slices one at a time without actually cutting the brain into extraordinarily thin slices. Taking images from multiple angles, Harris says, would allow researchers to reconstruct 3D structures and tease apart the mess.
The Allen Institute, by contrast, is using transmission electron microscopy (TEM) to build mouse connectomes. Reid says that his team has successfully reconstructed 1 cubic millimeter of visual cortex in the mouse using this technique, which is far cheaper than SEM but requires the brain to be sliced into ultrathin sections. Thus far, the different EM methods appear to have complementary uses. “We thought by this point one technology would win,” Reid says. “It’s nice to have more than one way to do something.”
The final step in the process will be to piece all the data together. First, a computer will stitch the images together, like the panorama function in a smartphone. Next, the computer will have to figure out which wires are which as they extend throughout the brain. For the mouse connectome, this might involve up to an exabyte of data — far too much for humans to process, says Viren Jain, a research scientist at Google in Mountain View, California. He says that the connectome team will most likely have to first use human volunteers or employees to identify some of these axons and how they connect to one another, then train a computer to do the same using machine learning algorithms. By the time the biologists are ready for data processing, Jain says, the algorithms should be up to the task.
An exabyte of data is equivalent to 1 billion gigabytes. “That’s certainly a lot of data, but not something that’s never been seen before,” Jain says. “In biology, it’s never been seen before.”
But he thinks that the task is feasible. “Ten years ago, if you asked someone about this, there would have been very little interest in even having this kind of data,” he says. “The progress made with the fly has people more motivated.”
The final challenge will be presenting the data in such a way that scientists everywhere can use it. “At the end, we’re motivated by the last and most exciting stage of discovery,” Seung says. A petabyte of image data will have to be compressed enough to be accessible through a web browser, while retaining enough detail to be useful to researchers looking to answer a specific question about the brain and its cells without getting overwhelmed. “It’s a problem of scale,” Seung says.
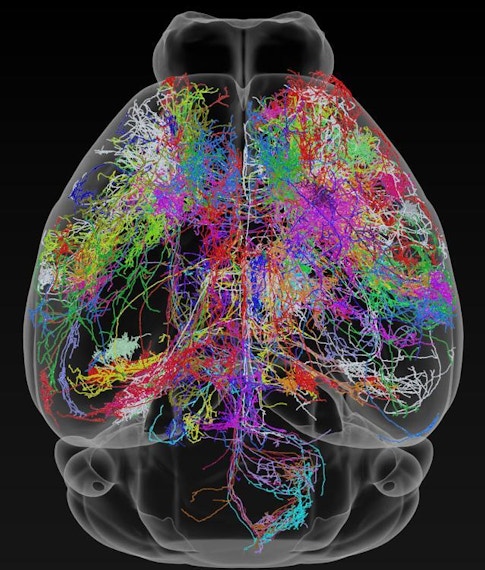
Building the best brain
Harris says that these engineering headaches should disappear once the first mouse connectome is finished, just as sequencing a human genome has become an everyday task that produces higher-quality data than the first human genome did two decades ago. “My suspicion is that the next connectome will be higher resolution,” she says.
Researchers are still debating how many mouse connectomes they should build. Some want to construct connectomes from multiple mice at once, arguing that each animal’s life and environmental exposure has altered its brain wiring in a different way. “You’ve captured one instant in its experience,” Ngai says. Evidence from C. elegans worms, whose 302 neurons have been extensively studied, suggests that the connections between these neurons vary greatly from one individual to another.
Others think that one connectome might be sufficient to at least provide the broad strokes of how a mouse brain works, even if the exact details differ from animal to animal. Once the engineering challenges are solved by the first connectome, the others could go more quickly.
“I’m looking forward to the time this’ll be routine,” Seung says.
Opinions also differ on which lucky rodent should provide the first brain connectome. Would it be more useful to reconstruct the brain of an inbred lab mouse, whose genetics and behavior are well studied, or the brain of a wild mouse which may be more intelligent and exposed to more stimuli throughout its life? Should the first mouse connectome be male or female, and how old should it be? Should it be genetically modified, and if so, which modifications would be the most informative?
These and other questions will be hashed out at the NIH/DOE workshops, which Harris is co-chairing, as they build toward a consensus. Ngai suspects that more than one brain will be necessary to elucidate phenomena like disease susceptibility. “We’ve already seen that from genetic analysis,” he says — white people in developed countries comprise the majority of fully sequenced genomes, and many findings in this group do not generalize to the human population as a whole. “By limiting ourselves to one homogenous population, these [predictions] fail.”
Behind all these discussions looms the intimidating next step: a human connectome. Given that the human brain is around 2,000 times the size of the mouse brain, the data contained in its wiring would number in the zettabytes. That’s approximately the amount of data generated in the world per year. “With EM, don’t hold your breath,” Reid says. “It’ll be several decades because it’s immense.”
Still, Reid expects scaling from mice to humans or another primate will be much easier than scaling from flies to mice. For one thing, the mouse connectome could provide some shortcuts, laying out the rules for how brain wiring works in mammals and eliminating the need to map every synapse. Lichtman likens these patterns to a chess game. Even though no two games are alike, chess follows a set of strict rules and predictable sequences. “It’s not a problem once you learn the rules; it’s all variations on a theme,” he says. “I think doing a whole mouse brain is going to teach us whether it makes sense to keep doing this for bigger and bigger brains.”
But Lichtman concedes that comparing connectomes may be necessary to explain why emergent properties like intelligence and consciousness differ so greatly between mice and humans. “We’ll need to do something beyond rodents to understand why rodents are not controlling the world,” he says.
The shortcuts provided by the mouse connectome may allow scientists to more precisely focus their efforts when building primate and human brain connectomes. In addition, a complete mouse connectome could allow scientists to test existing theories in a relatively simple system before wasting time and money testing incorrect theories in a complex one, bolstering efforts by theorists and experimentalists alike to build a detailed understanding of brain form and function.
“What’s exciting about connectomics is instead of waving your hand and thinking this circuit is how the brain computes, now you get the actual circuitry behind it,” says Hayworth. “The data you get really puts to the test the theories that are out there in a way that’s kind of brutal.”


