
How BrainCogs Learned to Share: A Case Study
Part 3 in the special report “The Data-Sharing Problem in Neuroscience.” Part 1 lays out the problems; part 2 describes some solutions.
When Manuel Schottdorf first arrived at Princeton in August of last year to begin his postdoc in David Tank’s lab, he was surprised to find that a large portion of the lab’s data was stored in immense Google spreadsheets housing millions of entries. Coming from the world of physics, Schottdorf was surprised at the lack of a more formal data infrastructure. The system worked well enough for individual researchers. But the lab had recently won a large-scale collaborative NIH grant with seven other groups, called BrainCogs, aimed at deciphering how the brain makes decisions based on working memory. These computations span the brain, so the various member labs, which each have expertise in different brain regions, would need to pool their data. “That data is so large, there is no way the experimenter who collected it can carry out all the analyses,” says Carlos Brody, a neuroscientist at Princeton and an investigator with BrainCogs and the Simons Collaboration on the Global Brain (SCGB). “Sharing the data so that others can work on it will be very valuable.”
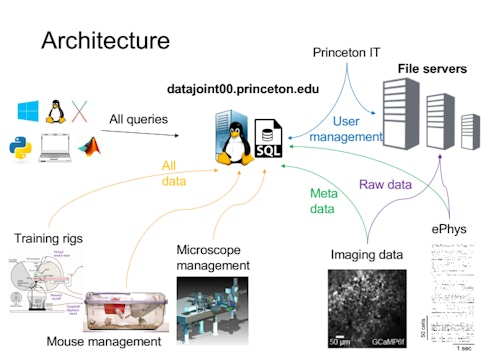
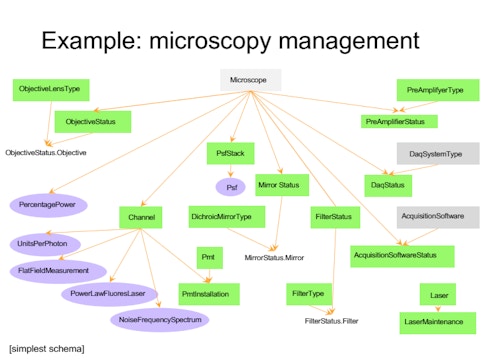
Each group has their own methods for storing data, which come with their own idiosyncrasies — one might group data by animal, another by day or experiment. “If you want to look across a dataset, you have to talk to each person to find out how they organize it, which takes a lot of time and effort,” Brody says. To develop a more robust system for organizing and sharing data, BrainCogs labs are adopting DataJoint, a tool for creating and managing scientific pipelines. “DataJoint helps you put a lot of that data in a database,” Brody says. “Then if you want to access it by day or by mouse ID or task, DataJoint will figure out how to access it and get that data.”
BrainCogs labs often study mice navigating a virtual maze. If all member labs have the data recorded during those experiments organized in a common format, querying the data as a whole is easier. For example, a recent visitor to the lab asked how task performance varies by time of day. Because researchers had transferred animal husbandry and behavior data to DataJoint, they could search the dataset and generate an answer in just 10 minutes. Before DataJoint, this would require writing code to compile everyone’s individual file formats. “Perhaps one of the biggest things this common framework gives us is that it’s much easier to do meta-analysis,” Brody says. “I don’t know if it’s the ideal solution, but it’s certainly helping standardize data formats and analysis pipelines and make it easier to share across people.”
BrainCogs has used some of its funds to support documentation of DataJoint’s tools, which researchers hope will help the community more broadly. Information on BrainCogs’ data pipeline is available on GitHub.
Despite the advantages, convincing everyone in BrainCogs to adopt DataJoint has been a challenge. “It’s not yet been adopted wholesale in our groups, but use is definitely growing,” Brody says. “People who already had existing data analysis code, which takes a long time to write, didn’t want to take the time to re-implement it in a way that’s compatible with DataJoint.” The most successful approach has been to start small, such as with animal husbandry. “That makes it easy for people doing it to get familiar with DataJoint and go from there,” Brody says. “We think that over time, it will become dominant framework,” he says.
Schottdorf and others are now working on a DataJoint pipeline for calcium-imaging data, which has been challenging. The datasets are huge, on the order of hundreds of terabytes, and each lab has its own approach for processing raw video data. BrainCogs groups will have to decide on a standardized approach, as will other U19 projects grappling with this issue. Schottdorf meets regularly with other researchers running the U19 data science cores to compare notes. He predicts that a core group will adopt a more or less standardized calcium imaging pipeline that will slowly spread. “But it’s not obvious how fast it will spread or how well it will work,” he says.
Standardizing Data Across Species: A Case Study
A multilab collaboration at the University of Washington is developing ways to compare data from humans and monkeys.


