
Taming the Wild Data World of Neuroscience
Part 2 in the special report “The Data-Sharing Problem in Neuroscience.” Part 1 lays out the problem.
Historically, neurophysiologists have been an individualist bunch. They often built their own rigs for probing the depths of the brain, and as technology advanced, wrote their own programs for handling data. This so-called gradware (for graduate student-written code) works for the person who created it. But it typically lacks the documentation and ease of use that accompanies professionally developed software, making it difficult to share. Without the person who wrote the code, the data may be largely inaccessible. “If data is only usable until the paper is published and the person leaves the lab, that data dies in a way,” says Carlos Brody, a neuroscientist at Princeton and an investigator with the Simons Collaboration on the Global Brain (SCGB).
As neuroscientists engage in larger collaborations, they are recognizing the need to develop more broadly useful and easy-to-use tools, a task that requires input from professional programmers. “High-quality software should not be built by grad students or contractors who are disconnected from the data and who don’t understand the science,” says Joshua Vogelstein, a neuroscientist at Johns Hopkins University. “The most successful strategy is to hire software engineers into academia that are excited about the science.”
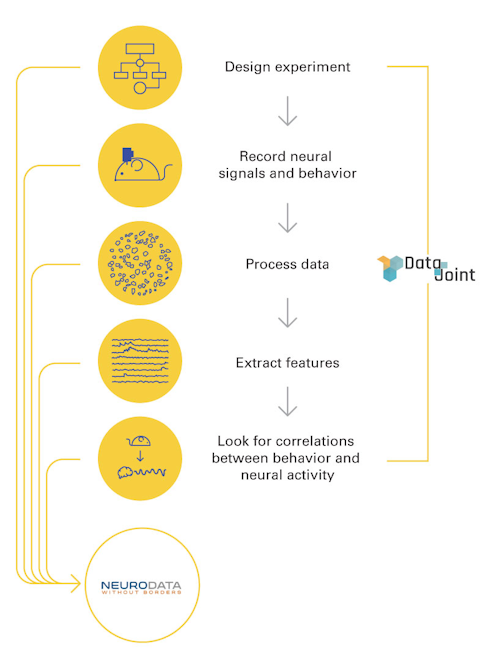
An emerging data standard called Neurodata Without Borders (NWB) is one such example. First launched in 2014, NWB was inspired by the emergence of several large-scale neuroscience projects, including Europe’s Human Brain Project, the Allen Institute’s MindScope and the BRAIN Initiative. It involved a close collaboration between neuroscientists and programmers from the get-go.
NWB acts as a sort of data tree that outlines how and where to store different types of data. “Properly documenting data so that someone outside the lab can understand it is one of the main goals of NWB,” says Ben Dichter, a data scientist who has been working with different groups to implement NWB and to develop new tools. “For example, if you want to store voltage recordings from implanted electrodes, you also have to specify the sampling rate. NWB tells you what to do so that you end up with the necessary metadata that you would need to start analyzing it.”
Over the last two years, NWB researchers and programmers from Lawrence Berkeley National Laboratory, led by Oliver Rübel, have expanded the platform, making it more flexible and modular. The newest version, NWB:N 2.0 (N for neurophysiology), was launched in January. NWB:N is designed to work with a variety of neurophysiology data, including electrophysiology, optical physiology and tracking and stimulus data.
The NWB team is working with the community to continue to improve the platform, such as enhancing compatibility with Matlab and Python and making it easier to convert data from different acquisition systems. As part of an SCGB-funded project, for example, the NWB team is working with Lisa Giocomo’s lab at Stanford to convert data from Neuropixels probes, a recording technology with hundreds of electrodes in a single probe. “People are really excited about this technology, and I think they will be willing to think about adopting a new data format as well,” Dichter says. Once developed, converters for Neuropixels and other data types will be freely available.
The NWB team is also building an ecosystem of analysis tools around NWB, such as calcium imaging tools and spike sorters. This has multiple advantages, including reducing duplicative efforts — a number of groups are developing similar calcium imaging tools, for example — and enhancing interoperability of different tools. “We really want converting to NWB to help with science today,” Dichter says. “It’s about creating an ecosystem where data and tools can be shared freely. We will end up with much healthier neuroscience community and be able to benefit from each other’s work much more.”
NWB held a hackathon at the Janelia Research Campus, in Reston, Virginia, in May to help researchers learn to use the software and to develop new tools. Participants included both users and developers, who worked on integrating NWB:N with data analysis, visualization, storage and other tools. Users arrived knowing very little about the platform and were able to transform their data into NWB format. “I think it was the most productive four days of my life,” says Kelson Shilling-Scrivo, a graduate student at the University of Maryland. “It’s nice when you have a problem with a code base to be able to talk to the guy who wrote it.” The NWB team also got feedback from users on the types of tools the community needs most.
A number of other ongoing efforts are converting existing data into NWB or developing new tools for NWB, which will then be available to the community. Allen and Berkeley Lab have a grant from BRAIN to further develop NWB using four specific use cases, each involving rodent behavior experiments ranging from visual detection to decision-making and navigation. Lydia Ng, senior director of technology at the Allen Institute in Seattle, is working on an NWB ontology, a formal delineation and definition of different properties and categories and the relationships between them. In addition, the Kavli Foundation offers $10,000 seed grants for labs interested in converting their data.
Support from prominent neuroscientists has been an important driver for NWB, as has interest from young researchers who have yet to become entrenched with other tools. “We’re seeing established neuroscientists willing to put themselves out there by sharing their data and tools,” Dichter says. “Simultaneously, new people coming into neuroscience are willing to think about the community in a whole new way.”
NWB offers the field a standardized data format, but that’s just one component of a data-sharing infrastructure. Researchers also want to be able to share their analysis pipeline — how they processed the data to get to the end result. This can be just as important as sharing the data itself, Brody says. “You can hand someone your hard drive, but that doesn’t mean they understand how you got to the figures in your paper.”
One such tool rapidly gaining traction among the neurophysiology community is DataJoint, a tool for creating and managing scientific pipelines originally developed in Andreas Tolias’ lab at Baylor College of Medicine that offers a way to organize disparate stores of data. “We believe the greatest obstacle to data sharing between collaborations and with the broader community is a deficit of structure and organizational consistency within projects,” says Dimitri Yatsenko, a research scientist at Baylor who helped develop and commercialize DataJoint. “A lot of effort is focused on organizing data after the fact. But we address organization of data within the active phase and teach people principles of data consistency and efficiency.”
Relational databases can be tricky to learn, but a benefit of DataJoint is that it can automatically translate Python and Matlab. “You get all the benefit of a relational database without the complications,” says Manuel Schottdorf, a postdoctoral researcher at Princeton University who oversees data sharing for a multi-lab project. “With a unified data structure and tools, we can be more productive because we don’t have to reinvent the wheel.”
DataJoint is free and open source — labs can adopt it on their own or contract with DataJoint for additional support. “We strive to provide sufficient documentation and resources for labs to use DataJoint independently, and as a company, provide support for groups that need additional help,” says Yatsenko, who provides consulting services to science labs to configure their data processing pipelines. “But the tools are designed to be learned and used by the researchers themselves.”
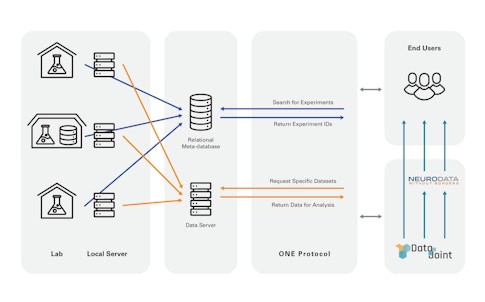
DataJoint’s largest ongoing project is with the International Brain Lab, a multi-lab collaboration that must share data among its 21 member labs. DataJoint helped the IBL set up a data pipeline in the cloud, providing tutorials for member labs to learn to use it. Researchers can query specific types of data through the pipeline, which makes it less unwieldy, Yatsenko says. “You can get the exact chunk of data you want to your laptop instead of large datasets.” The pipeline can also be configured to automatically carry out certain computations as soon as the data is available upstream. This is useful for quickly assessing data quality, for example, an important metric for IBL labs. “DataJoint doesn’t give a fixed pipeline, but gives a language and tools to create a pipeline with data integrity at each step,” Yatsenko says.
NWB and DataJoint are working to make their systems function together, developing tools to read NWB data into a DataJoint pipeline and to convert data from DataJoint into NWB.
—
How BrainCogs Learned to Share: A Case Study
Members of the multi-lab BrainCogs collaboration are using DataJoint to more easily access each other’s data.
Standardizing Data across Species: A Case Study
A multi-lab collaboration at the University of Washington is developing ways to compare data from humans and monkeys.
—
It’s not yet clear which standards will win out in neuroscience. Finding an infrastructure with the flexibility to suit the field’s diverse needs and evolving tools, as well as the robustness to stand up over time and a low enough barrier to entry to encourage broad adoption, is a tricky balance. “Standards are challenging because they necessarily involve compromises,” Dichter says. “But they also facilitate the next level of innovation. You can focus energy on actual scientific problems and more efficiently iterate through different types of analysis.”
Despite the clear advantages of a data standard, Kenneth Harris, a neuroscientist at University College London and part of the IBL, cautions against settling down too soon. He instead supports an organic rise to the top — a period of experimentation and evolution followed by a community-driven consensus on the optimal format. “I think it’s important not to put all our eggs in one basket, or even two or three,” Harris says. “In other fields, it took a few rounds of improvements to get things sorted out, but within a few years of large-scale projects starting, that’s when standardization happened.”
But he echoes Dichter’s excitement about the science that standardization efforts will enable. “Building data architecture is not why I got into science, but it has to be done,” he says. “Soon we will be able to figure out things we couldn’t do before because we didn’t have infrastructure.”
Neuroscience data is highly diverse, making it difficult to create tools for data-sharing and standardization.


