New Neuronal Mapping Technique Reveals Surprising Cortical Connections
Mapping how information flows through the brain is a serious challenge. Even the diminutive mouse brain has 75 million neurons — and orders of magnitude more synapses — wrapped into a dense tangle of tissue. “We don’t really understand how information is routed in the cortex,” says Anthony Zador, a neuroscientist at Cold Spring Harbor Laboratory in New York and an investigator with the Simons Collaboration on the Global Brain (SCGB).
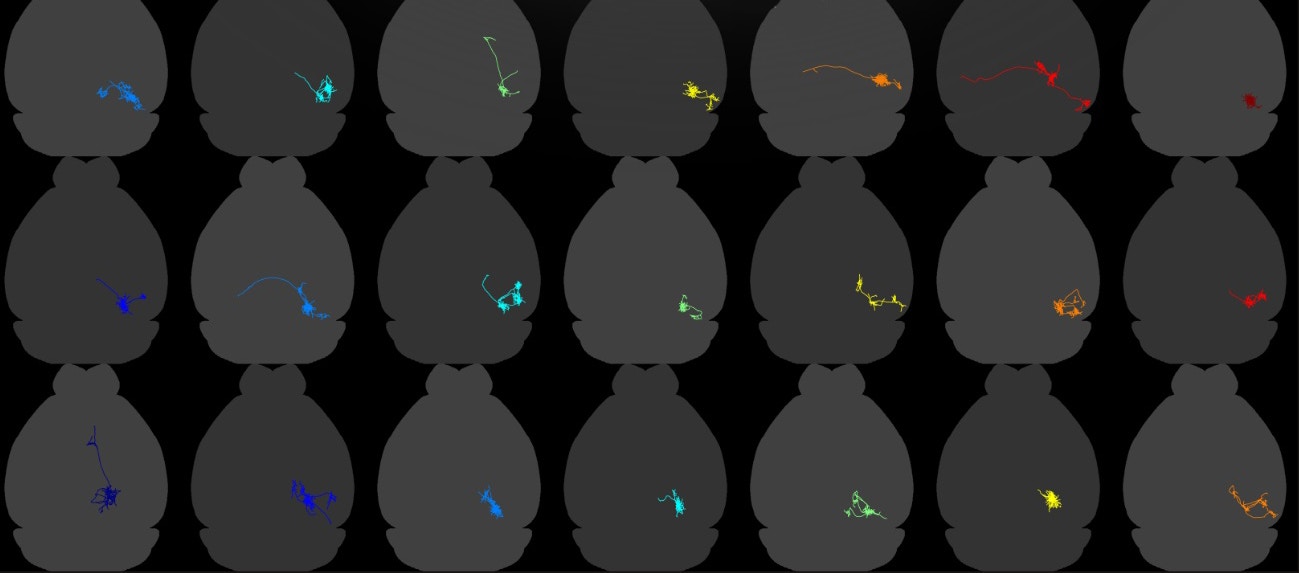
A new technique called MAPSeq, developed by Zador and his collaborators, uses genetic barcodes to label cells, making it possible to map hundreds or potentially thousands of individual neurons within this tangle. In a study published in Nature in March, scientists used this approach to efficiently trace hundreds of cells in the visual cortex.
The effort revealed that single cells in this region project to two to seven different downstream regions, a more distributed pattern than scientists expected. And these connections are not random, suggesting there is a higher-order structure in how neurons choose to project.

The findings challenge the idea that the visual system has dedicated pathways for coding certain features. “In the mouse visual cortex, neurons send information to several different places rather than to dedicated channels,” says Thomas Mrsic-Flogel, director of the Sainsbury Wellcome Centre in London and an SCGB investigator.
Previous efforts to trace neuronal projections have examined handfuls of cells in specific parts of the brain. But none had looked systematically at large numbers of neurons in a single area. “We had no idea how many areas individual neurons in the neocortex project to,” Mrsic-Flogel says. That kind of information is important for understanding how information is conveyed to downstream structures, he says.
Indeed, very little work has been done on what Mrsic-Flogel dubs the cell’s projective field — how a neuron influences downstream neurons in the network. By contrast, neuroscientists have extensively mapped neurons’ receptive fields, the specific features in an image or other stimuli that activate a particular cell.
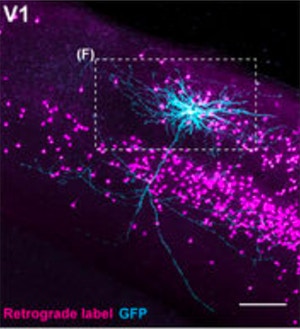
In the new study, researchers used two different methods for tracing neuronal projections. First, they labeled individual cells in the visual cortex with a fluorescent tracer and used two-photon imaging to trace where the cells sent their axons. All of the several dozen individual neurons they analyzed sent projections outside of the visual cortex, most to two or more areas. “Each neuron engages in both local and distal computations,” Mrsic-Flogel says.
MAPseq analysis of more than 500 visual cortex neurons revealed that projections were not random but followed a higher-order logic. Most neurons preferentially projected to specific combinations of areas. In some cases, they even avoided certain areas.
Zador says he was somewhat surprised by the result. He expected that neurons in the visual cortex would follow a few specific projection patterns — some might project to areas A and B, some to C and D, and so forth. But instead, the patterns are highly diverse. “We are still trying to make sense of what that structure means,” he says.
Based on this and other research, Zador hypothesizes that the brain dynamically changes how it routes information, perhaps choosing subsets of targets based on what the animal is doing. For example, when a mouse is navigating a maze, it may send visual information to areas A, B and C. But when it’s foraging for food, visual information might need to go to B and D.
Zador and his collaborators have developed a new iteration of MAPSeq called BARseq that allows researchers to map multiple measures in the same neurons: the bar codes, neuron activity via calcium imaging and gene-expression data. (For more on MAPseq and BARseq, see ‘RNA barcodes for tracing neurons.’)
Now that researchers have a way to create more detailed maps of neuronal projections, they can ask more sophisticated questions about how information flows through the system. For example, do neurons that target two specific areas encode different information than those targeting other areas? Do neurons that innervate multiple areas carry more general information than those that target one area? Do neurons that target two different areas help to coordinate activity in those areas, influencing how that information is processed? Mrsic-Flogel, Zador and their collaborators aim to tackle these and other questions in the International Brain Lab, a collaboration among 20 different labs funded by the SCGB.


