Using Artificial Intelligence to Map the Brain’s Wiring
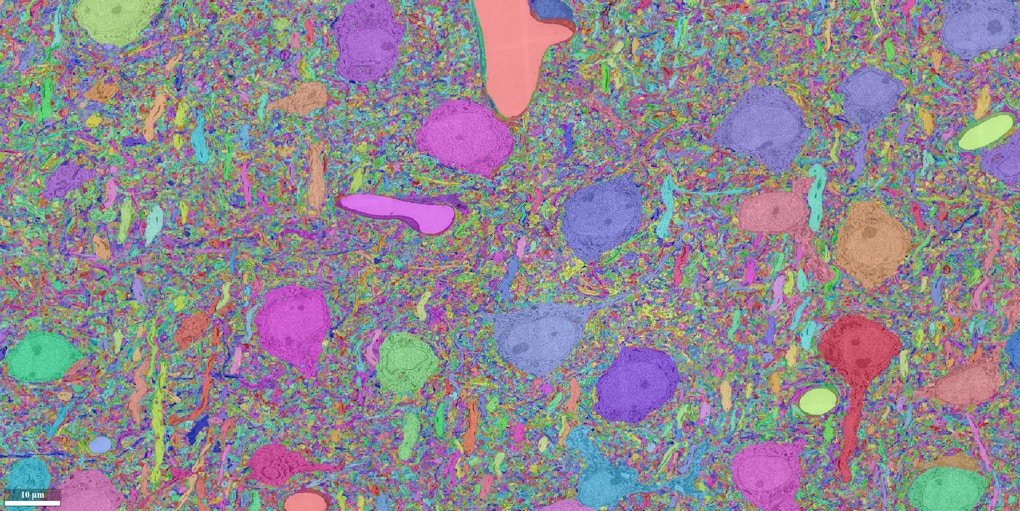
In July, neuroscientist Sebastian Seung and his collaborators finished an 18-month marathon task. Working with roughly a terabyte of data, his team used artificial intelligence to reconstruct all the neural wiring within a 0.001 cubic millimeter chunk of mouse cortex. The result is an intricate wiring diagram that Seung and others will mine for insights into the cortex, such as how the circuits are connected and how they perform computations.
Seung hasn’t had time to savor the accomplishment, however. His team is racing to complete the next stage of the project — they plan to expand their efforts a thousand-fold to reconstruct a cubic millimeter chunk of mouse cortex. Both maps are part of a multi-lab program called Machine Intelligence from Cortical Networks (MICrONS), which is funded by Intelligence Advanced Research Projects Activity (IARPA), the U.S. federal government’s intelligence research agency. The project aims to improve artificial intelligence by enhancing our understanding of the brain. “The neuroscience goal of the five-year program is to observe the activity and connectivity of every neuron in a cubic millimeter of mouse neocortex,” Seung says.
Seung’s IARPA team includes labs at Baylor College of Medicine in Houston, Texas, and the Allen Institute in Seattle, Washington. The Baylor group is focused on physiology, using calcium imaging to track neural activity within the target cube of brain. They’ll then ship the tissue to scientists at the Allen Institute, who will perform scanning electron microscopy of the same cubic millimeter. The Allen group will send the resulting data to Seung’s lab, which will reconstruct the connections among 100,000 neurons. Two other IARPA-funded teams are attempting the same feat in parallel, using different techniques.
Seung’s machine-learning algorithms are an essential component of the program. Without artificial intelligence (AI), the first phase of the project — reconstructing the 0.001 cubic millimeter piece of mouse cortex — would have required roughly 100,000 person-hours. Seung says their success lay in part in enormous advances in machine learning that have unfolded over the past five years. “We surprised even ourselves by how much we could improve the accuracy of artificial intelligence in less than 18 months,” Seung says.
From gaming to the brain:
Seung’s foray into connectomics began in 2006, when the neuroscientist decided to dramatically shift the focus of his lab. Scientists were building tools to image the brain at synapse-level resolution, but stitching together reams of microscopy data was prohibitively slow. The field needed an efficient alternative. Seung decided to apply a brain-inspired machine-learning technique he had encountered when working at Bell Labs in the 1990s — convolutional neural networks.
To create neural wiring diagrams by hand, scientists start with cross-sectional images of a slice of tissue. Each image features cross sections of individual axons and dendrites, which the scientist traces and tracks through the stack of images. The end result is a painstaking reconstruction of the cells and their many connections. Machine-learning algorithms take a similar approach, using a hand-labeled data set to learn to trace the boundaries of neurons and connect them into a cohesive wiring map. “The computer learns to emulate the human judgments,” Seung says.
Seung’s early algorithms helped to automate the reconstruction process, but the effort still needed human input. So in 2012, Seung and his collaborators launched a crowd-sourced game called Eyewire, in which players could trace neural wiring on a computer.
An army of more than 100,000 ‘citizen neuroscientists’ have played the game, generating a neural dataset that has culminated in two publications to date. Researchers used the data to identify a new cell type in the retina and to show how some retinal neurons respond to direction of movement. (The ‘Eyewirers’ are listed as authors on both papers.) Images of reconstructed neurons from Eyewire are available at the online Eyewire Museum.
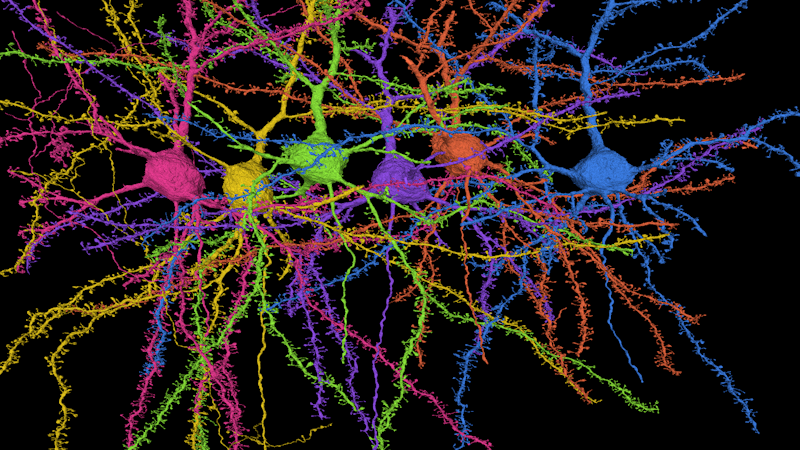
A reconstruction of six pyramidal cells, the most popular type of cell in the cortex. The cells’ dendrites are covered in tiny spines, the site of synaptic connections. Credit: Seung lab
Around the same time that Eyewire was taking off, the field of deep learning was undergoing a revolution. The development of graphics processing units, or GPUs, for video games provided a powerful computing platform to execute neural network algorithms that had been developed years earlier. “GPUs were built so that 10-year-old kids could play video games, but they just so happen to be great for AI,” Seung says.
With this new technology in place, neural network algorithms advanced rapidly, evolving into the image- and voice-recognition software that has become ubiquitous on smartphones and other devices. Seung and his collaborators applied the same advances to their connectomics software, enhancing its ability to reconstruct the brain. “We are being carried along by the deep-learning tsunami,” Seung says. “We benefit from the efforts of an entire community and investment of billions from famous companies.”
The newest versions of the software are faster and more sophisticated, capable of distinguishing between axons and dendrites, for example. Indeed, Seung’s team’s newest software is so good it can sometimes outcompete its human counterparts. In 2013, the researchers developed a machine-learning challenge to reconstruct 3-D electron microscopy data. Participants trained algorithms on a labeled dataset and then tested their contenders on two novel sets of images. Seung and others have continued to refine and test their algorithms, comparing their performance to the two human scorers who originally annotated the data.
Earlier this year, one of the algorithms beat the human version. “With a lot of caveats, this is the first time we have a computer with superhuman accuracy,” Seung says. “One could argue that the means of generating the ground truth is not good. But for many years, no one could beat the estimate of human accuracy.”
1,000-fold improvement
Despite these rapid advances in AI, the next phase of the IARPA project is daunting. Reconstructing a 1,000-fold larger volume of brain tissue will require a 1,000-fold improvement in AI, or a combination of AI and human supervision. The dataset will grow from terascale to petascale, which will likely require new systems to manage and process the flood of information.
Seung is confident that his team is up to the task. “Because of advances in deep learning, we have great potential to be able to analyze that data in a reasonable time,” Seung says. “I think people feel more optimistic than ever that fully automated reconstruction with almost no human effort will be feasible. We aren’t there yet, but it will be feasible.”
In the meantime, human effort, provided in part via crowdsourcing, will provide an important component of the project. “Even if we improve AI by a factor of 100, we still need another 10-fold,” Seung says. “We want to mobilize people to help us.”
To that end, the researchers are working on the successor to Eyewire, called NEO. “We’ve made a quantum leap in AI, which requires different kinds of crowdsourcing platforms and different ways for humans to interact with AI,” Seung says.
Whereas the efforts of Seung’s lab largely focus on connectivity, the MICrONS project is bringing together both anatomy and physiology. “I think that the combination of anatomical and physiological information at this large scale is going to really accelerate our understanding,” Seung says. For example, he says, having a connectome map may influence how neuroscientists design their physiology experiments.
Seung hopes that the technologies they develop for the IARPA project, including automated image acquisition and automated analysis and reconstruction of circuits, will spread much more broadly. “I think as the technology develops, it will be easy for anyone to adopt,” he says.
Seung’s efforts have relied heavily on advances in AI. Eventually, IARPA aims to close the loop, using new insights into the brain to further enhance AI. Neuroscience inspired the first generation of artificial neural networks, and now neuroscientists are using these networks to understand the brain, Seung says. “There’s no reason not to go around the loop more than once.”
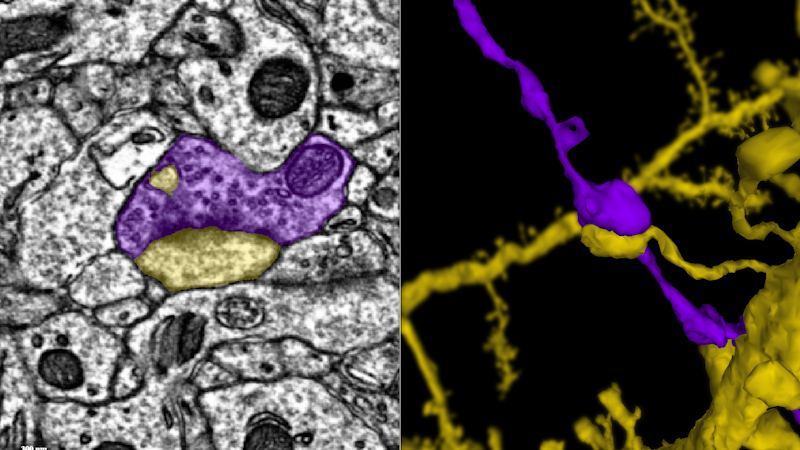
This movie follows a single axon (blue) until it makes a synapse onto the dendrite of another neuron (green). Credit: Davit Buniatyan & Nico Kemnitz, Seung Lab